A frequent image analysis technique that is performed routinely at Visikol is the colocalization of many different biological labels in fluorescent images. A colocalization analysis is the identification of spatial overlap between multiple labels. This type of analysis is important in identifying areas where two or more labels are strongly expressed, which can give valuable information about the state of each cell in the image.
This image analysis can also give researchers information about where certain markers are localized and can identify cells of differing phenotypes. It is important to know how to perform this type of analysis so that one can gleam important information from their images. Therefore, the information below is a simple run-through of a basic method to colocalize two labels utilizing ImageJ. At Visikol, we use various image analysis packages such as HALO from Indica Labs for image analysis but also routinely use ImageJ due to how well validated and used it is in the literature.
PART 1: Preparing the Images
Once the labels of interest have been imaged, open two images to colocalize in ImageJ (make sure each image contains only one label and that each image is the same size and of the same region), then threshold the images to obtain a mask of the labels. Depending on the thresholding, use various filters to de-noise the image to clean it up. An example is shown in Figure 1 and Figure 2 of the masks obtained from thresholding images of DAPI and Phalloidin using the OTSU auto-threshold method.
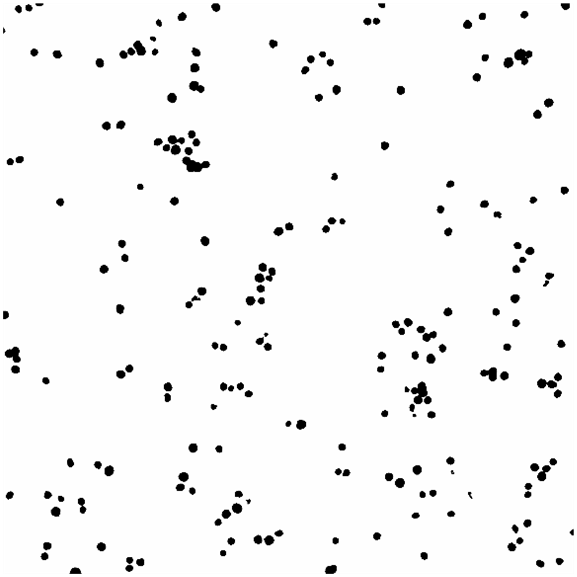
Figure 1. Images of the masks for DAPI
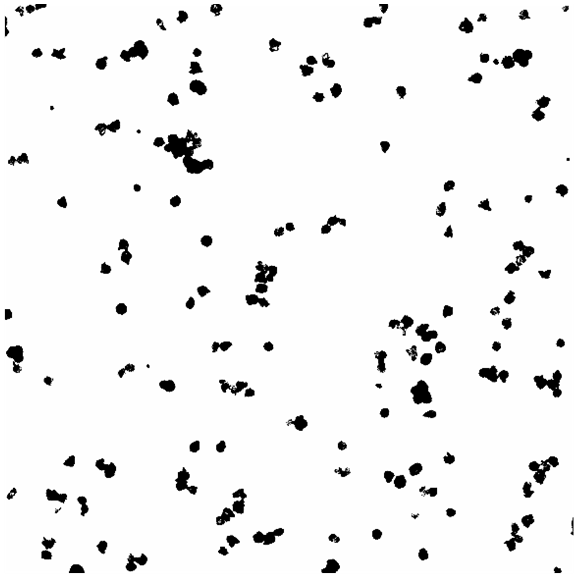
Figure 2. Image of masks for Phalloidin.
PART 2: Colocalization of the Masks
In order to colocalize the images, use the image calculator under the process tab of ImageJ. Once the window pops up, input the images for colocalization and use the “AND” operation, as in shown in Figure 3 below. The “AND” operation works by taking the two input images and extracting all the pixels where each label overlaps in order to form a new image. The new image formed is a mask of the colocalized regions of the two labels. An example can be seen in Figure 4 below.
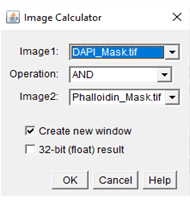
Figure 3. An example image of the image calculator window for ImageJ and what inputs to use.
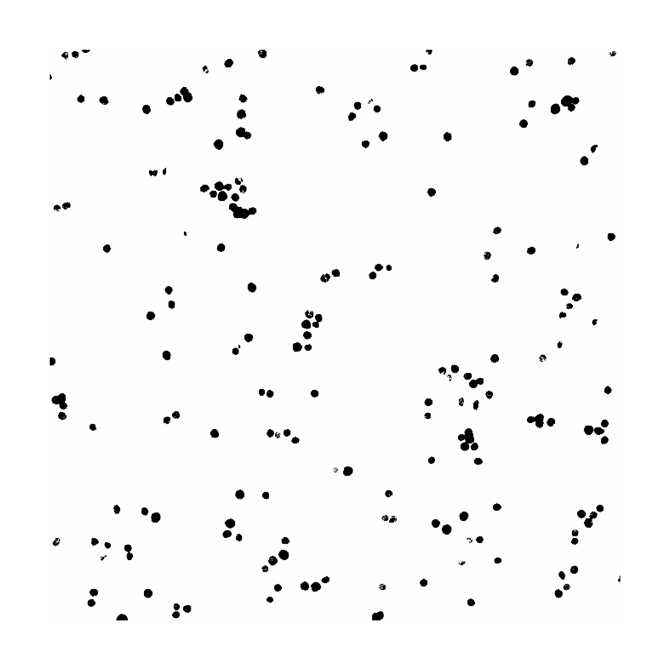
Figure 4. The colocalized image of DAPI and Phalloidin
Colocalization is an important type of analysis that researchers can use to visualize where labels overlap, which can give vital information about the image in question, and the method shown above is very basic. Visikol performs more advanced colocalizations quite frequently and has methods to colocalize any number of labels together and obtain object level data about each label in each cell in an image.
The more advanced colocalizations are very useful for figuring out cell phenotypes, regions of overlap, and region-specific information about various fluorescent samples. If you need an advanced colocalization done or have any questions about colocalization in general, please feel free to reach out to Visikol.
