Once you’ve imaged your well plates using a high content microscope, you’ll have a huge collections of images to process. ImageJ is a great tool for processing images, but when you have too many images to count, it can be extremely time consuming to process them manually. This blog post outlines a simple process for converting your image processing workflow into an automated macro that will process all the images in your well plate the same way, so you can get the analysis and interpretation of your results without the headache of manually processing thousands of images. In this blog post, you will learn how to use create an ImageJ macro that will use thresholding to detect cells in an image, and measure the area, intensity, and shape characteristics of each nucleus. It will output a data table with the measurements for each nucleus detected in a set of images from a well plate.
Step 1. Open one of the images from your image set
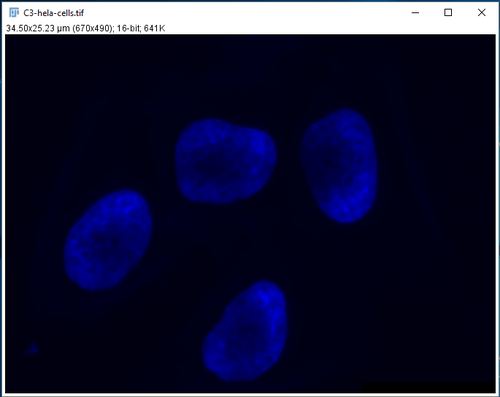
For this tutorial, we will be thresholding and analyzing the nuclear channel of HeLa cells, to count the number of cells and measure the area, mean intensity, and shape of each nucleus. A sample of an image of HeLa cells is available built-in to ImageJ, just click File > Open Samples > HeLa cells, and split the channels to leave the nuclear channel (channel 3).
Step 2. Record Macro Steps to be Performed on Each Image
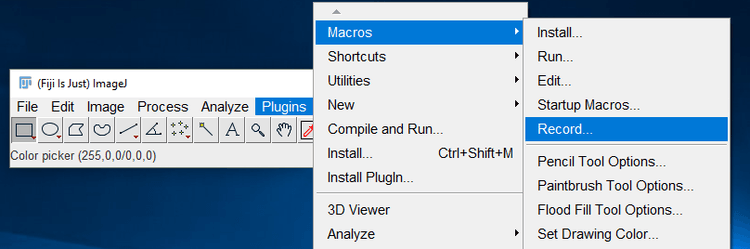
ImageJ comes with a built in tool for recording the steps you perform on an image into the ImageJ macro language. This is a convenient way to grab the actions you take so that they can be incorporated into a batch macro. To open the Macro recorder, click on the Plugins menu item > Macros > Record…
The Macro recorder window will appear. When this window is showing, any actions you take in ImageJ will be converted into macro code. This is very useful when creating a macro, as you can perform ImageJ functions manually, and the resulting macro code will be given to you. Then with slight modifications, the recorded code can be applied to large numbers of images.
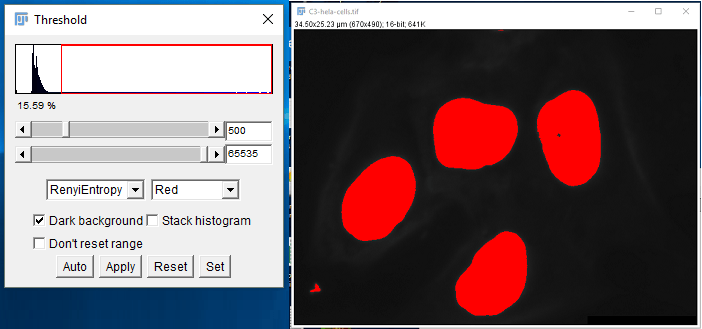
Now we are ready to record the macro. The first step will be to threshold the image. To open the Thresholding dialog, click the Image menu then select Adjust > Threshold (or pressing Ctrl+Shift+T). The Thresholding dialog will pop up. Since we are automating analysis, we want to select an automatic thresholding method. For this image set, the RenyiEntropy method works well. Make sure “Dark background” is selected, or else you will threshold the background instead of the foreground.
Next we need to select the measurements we want to obtain from the particles. On the ImageJ menu, select Analyze > Set Measurements. The Set Measurements dialog will pop up. Select Area, Mean gray value, Perimeter, and Shape descriptors (for circularity measurement). Also select Limit to threshold, otherwise ImageJ will calculate this measurements on the whole image instead of the thresholded areas, which is not what we want to do. Select OK.
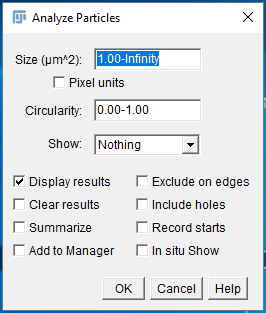
The next step is to use the Analyze Particles function in ImageJ to detect the cells and measure their characteristics. Click the Analyze menu, and then select Analyze Particles. The Analyze Particles dialog will pop up. For the size parameter, enter in 1.00-Infinity. We enter 1 um² as the lower limit, that way we will eliminate any specks of dust that show up in the image. Select “Display results” since we want to save the results to the results table after analysis. Click OK. The results table will pop up, showing you 4 rows, one row for each cell detected.
Now take a look at the Macro recording window. You should see several lines of code, corresponding to all the actions you’ve taken in ImageJ so far. Your code should look like this:
//run("Threshold..."); setAutoThreshold("RenyiEntropy dark"); run("Set Measurements...", "area mean perimeter shape limit redirect=None decimal=4"); run("Analyze Particles..."); run("Analyze Particles...", "size=1-Infinity circularity=0-1.00 display");
The first line is commented out (everything on a line “//” in ImageJ code is ignored, known as a comment), since that line would show the Threshold dialog box, and we want to set the threshold automatically, not show the dialog box for user input. Also notice that there are 2 lines which show the run(“Analyze Particles…”) command. The first line where this appears is not necessary, since that line will show the dialog box for the Analyze Particles window, so you can delete that line, since for automating the analysis of a whole plate, we don’t want to show the Analyze Particles dialog for every image. The second line actually calls the Analyze Particles command and gives it the parameters needed to execute the function, this is the line we need.
Step 3. Create the Macro
Now we are ready to get to the coding. To create a new macro, on the ImageJ menu, click Plugins > New > Macro. The Macro code box will pop up.
First, we will create a function to execute the steps we just recorded. We will call the function “processImage” and it will contain the lines that were recorded using the macro recorder.
function processImage(imageFile) { setAutoThreshold("RenyiEntropy dark"); run("Set Measurements...", "area mean perimeter shape limit redirect=None decimal=4"); run("Analyze Particles...", "size=1-Infinity circularity=0-1.00 display"); }
Now, we need to add some code. We need to add code to open the image first. Then we need to add code so that the filename of the image being processed is added to the results table. This way, you can break down the results by which well they came from later. Finally, we need to close the new images that were created.
function processImage(imageFile) { // Store the number of results before executing the commands, // so we can add the filename just to the new results prevNumResults = nResults; open(imageFile); // Get the filename from the title of the image that's open for adding to the results table // We do this instead of using the imageFile parameter so that the // directory path is not included on the table filename = getTitle(); setAutoThreshold("RenyiEntropy dark"); run("Set Measurements...", "area mean perimeter shape limit redirect=None decimal=4"); // You should modify the size parameter in the line below based // on the calibration of your images run("Analyze Particles...", "size=1-Infinity circularity=0-1.00 display"); // Now loop through each of the new results, and add the filename to the "Filename" column for (row = prevNumResults; row < nResults; row++) { setResult("Filename", row, filename); } close("*"); // Closes all images }
The next step is to add code to ask the user of the macro to select a directory of images to process, and then to call our processImages() function on each image in the directory, and finally to save the results. The code is shown below:
run("Clear Results"); // clear the results table of any previous measurements // The next line prevents ImageJ from showing the processing steps during // processing of a large number of images, speeding up the macro setBatchMode(true); // Show the user a dialog to select a directory of images inputDirectory = getDirectory("Choose a Directory of Images"); // Get the list of files from that directory // NOTE: if there are non-image files in this directory, it may cause the macro to crash fileList = getFileList(inputDirectory); for (i = 0; i < fileList.length; i++) { processImage(fileList[i]); } setBatchMode(false); // Now disable BatchMode since we are finished updateResults(); // Update the results table so it shows the filenames // Show a dialog to allow user to save the results file outputFile = File.openDialog("Save results file"); // Save the results data saveAs("results",outputFile);
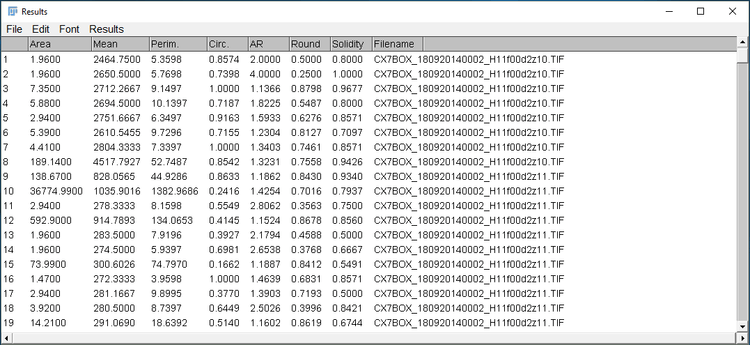
Now all the pieces of the macro are assembled, and it’s ready to run! Running this macro on a directory of images, we see the following results. It processed 350 images in a little over 1 second.
The full macro code is shown below. It can be copied and pasted directly into ImageJ, or you can download the macro file.
Full Macro Code:
// Macro to measure Area, Intensity, Perimeter, and Shape of directory of images run("Clear Results"); // clear the results table of any previous measurements // The next line prevents ImageJ from showing the processing steps during // processing of a large number of images, speeding up the macro setBatchMode(true); // Show the user a dialog to select a directory of images inputDirectory = getDirectory("Choose a Directory of Images"); // Get the list of files from that directory // NOTE: if there are non-image files in this directory, it may cause the macro to crash fileList = getFileList(inputDirectory); for (i = 0; i < fileList.length; i++) { processImage(fileList[i]); } setBatchMode(false); // Now disable BatchMode since we are finished updateResults(); // Update the results table so it shows the filenames // Show a dialog to allow user to save the results file outputFile = File.openDialog("Save results file"); // Save the results data saveAs("results",outputFile); function processImage(imageFile) { // Store the number of results before executing the commands, // so we can add the filename just to the new results prevNumResults = nResults; open(imageFile); // Get the filename from the title of the image that's open for adding to the results table // We do this instead of using the imageFile parameter so that the // directory path is not included on the table filename = getTitle(); setAutoThreshold("RenyiEntropy dark"); run("Set Measurements...", "area mean perimeter shape limit redirect=None decimal=4"); // You should modify the size parameter in the line below based // on the calibration of your images run("Analyze Particles...", "size=1-Infinity circularity=0-1.00 display"); // Now loop through each of the new results, and add the filename to the "Filename" column for (row = prevNumResults; row < nResults; row++) { setResult("Filename", row, filename); } close("*"); // Closes all images }