Overview
At Visikol, we specialize in advanced biological image analysis services, and one of our core offerings is Cell Population Analysis. Our comprehensive analysis techniques revolutionize the way you analyze histological sections, enabling accurate cell segmentation and precise quantification of marker positivity within cells.
Quantifying the total number of cells within histological sections is a crucial measure in various applications, but manual cell counting is laborious, error-prone, and subject to biases. Our computerized image analysis solution provides a reliable and efficient method for establishing accurate cell counts. Whether you need to quantify cells within tumor boundaries, assess hormone receptor positivity, or measure immune infiltration such as T-cells or polarized macrophages within a tumor, our services offer comprehensive assessments on histological sections.
Immunohistochemistry (IHC), whether chromogenic or fluorescence, is a widely used technique for determining cell populations, evaluating disease markers, and assessing the effects of drug treatments on tissues. Traditionally, the analysis of IHC slides relied on subjective evaluations by pathologists, often utilizing scoring systems to indicate the presence or absence of specific markers. However, with our modern computational techniques and advanced image processing capabilities, we can accomplish quantitative analysis on large numbers of tissue sections in a fraction of the time.
At Visikol, we offer a suite of services for the quantification and analysis of H&E and IHC sections, allowing you to examine the extent and distribution of disease-relevant cells with precision. Our automated image processing techniques enable the breakdown of subpopulations of cells, determination of coincidences and spatial relationships between cell types, and evaluation of protein expression changes in specific cell types, among many other possibilities.
Key Features of our Cell Population Analysis Service:
- Automated Cell Segmentation: Our advanced algorithms accurately segment cells within histological sections, eliminating the need for labor-intensive manual counting and minimizing errors and biases.
- Marker Positivity Quantification: We precisely quantify the level of marker positivity within individual cells, providing quantitative measurements of biomarker expression.
- Subpopulation Analysis: By analyzing the colocalization of different biomarkers within cells, we identify and quantify distinct subpopulations, allowing for a deeper understanding of cellular heterogeneity.
- Spatial Relationships: Our services evaluate the spatial relationships and coincidences between different cell types, providing insights into cellular interactions within tissues. See the Spatial Analysis page for more detail.
- Comprehensive Assessments: Whether you’re studying tumor microenvironments, evaluating disease markers, or assessing the effects of drug treatments, our services provide comprehensive and reliable analyses on histological sections.
Protocol
| File Formats | TIFF, PNG, JPEG, CZI, MCD, SCN |
| Image Submission | Hard Drive Visikol Cloud Sharing Client selected file sharing service |
| Data Delivery | Report Tabular data *Segmentation masks available upon request |
General Procedure
- Images are checked to make sure the quality of the images is good for analysis.
- Cells are segmented through a nuclear marker.
- The cytoplasm is extrapolated through expansion or with a cytoplasmic marker.
- The other markers are thresholded to determine positive areas within the images.
- Thresholded markers and raw markers are colocalized with the segmented cells to obtain positivity and intensity measurements.
- The organized data and report are then delivered to the client.
Representative Data
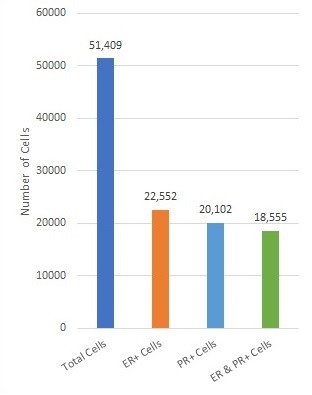
Quantification of Cells Expressing Hormone Receptors in Clinical Breast Cancer Biopsy Sections
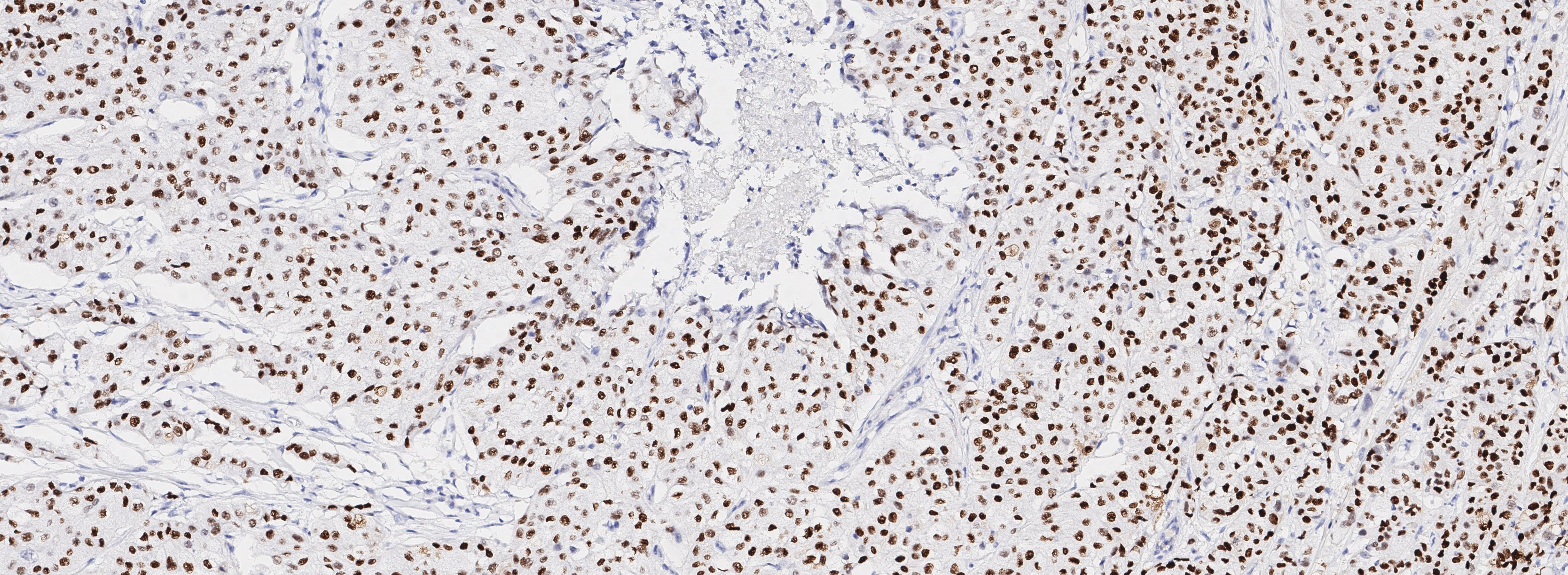
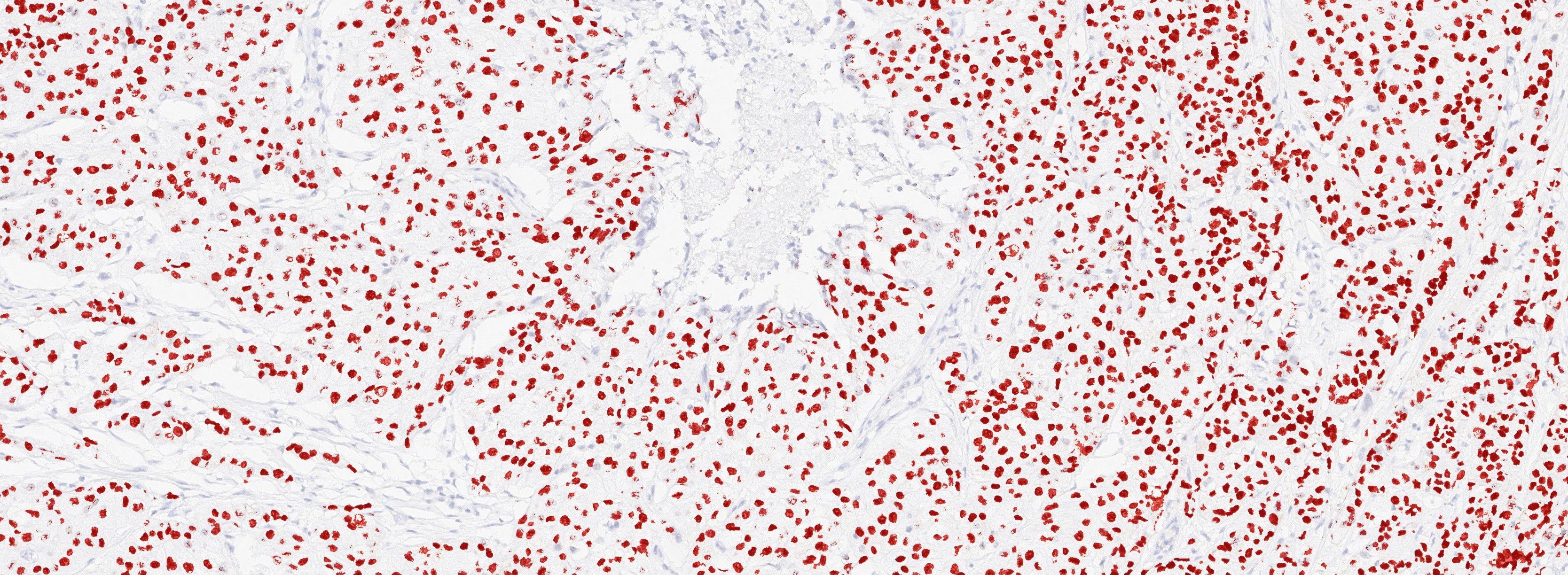
Figure 1A. Tissue section from biopsy of female patient diagnosed with invasive ductal adenocarcinoma, stained for estrogen receptor (ER); Move slider to reveal segmentation of ER+ cells, which are highlighted in red.
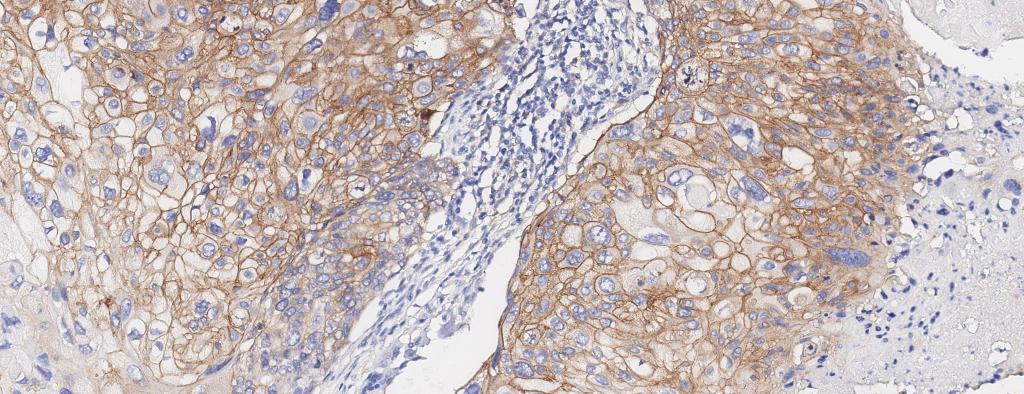
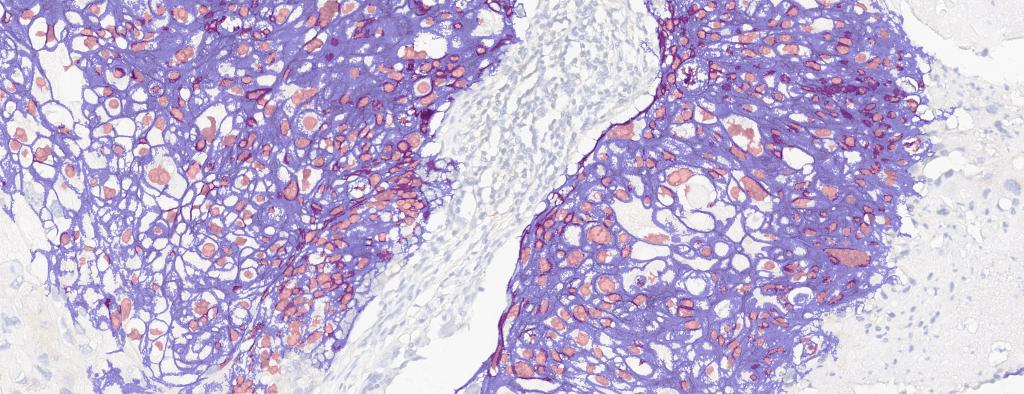
Figure 2. Breast cancer biopsy section from 50 y/o female patient with metastatic invasive ductal adenocarcinoma, stained for EGFR. EGFR+ cells identified (drag slider to see segmentation of EGFR+ cells)
Quantification of Astrocyte Reactivity and Spatial Distribution in TBI-induced Rat Brain Tissue
Investigation of the activation of astrocytes within rat brain following mechanically induced severe traumatic brain injury (TBI) showed a significant increase in reactive astrocytes. Quantification of two parameters, the distance between neurons and nearest reactive astrocyte, and distance of reactive astrocytes to nearest blood vessel was conducted on rat brain sections labeled with GFAP. The average neuron-astrocyte distance found in control samples was consistent with reported results [2]. After mechanically induced traumatic brain injury, the distance between neurons and activated astrocytes was found to decrease in a significant manner. The astrocyte-blood vessel distance was found to increase due to increases to astrocyte activation and GFAP expression following TBI. A statistically significant increase in GFAP expression leading to a significant increase in the number of astrocytes detected following TBI, consistent with previously reported results [3].
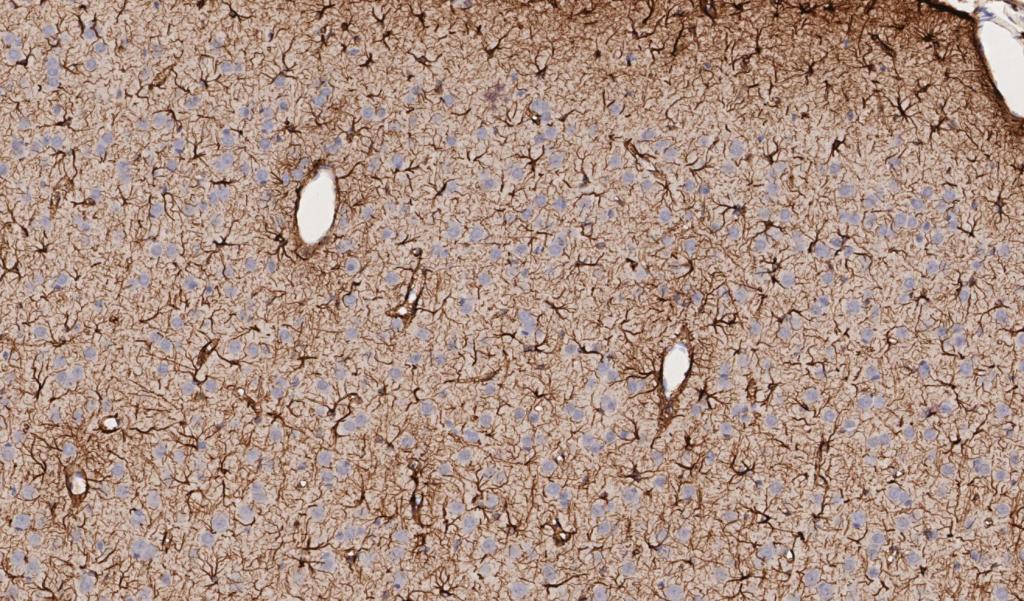
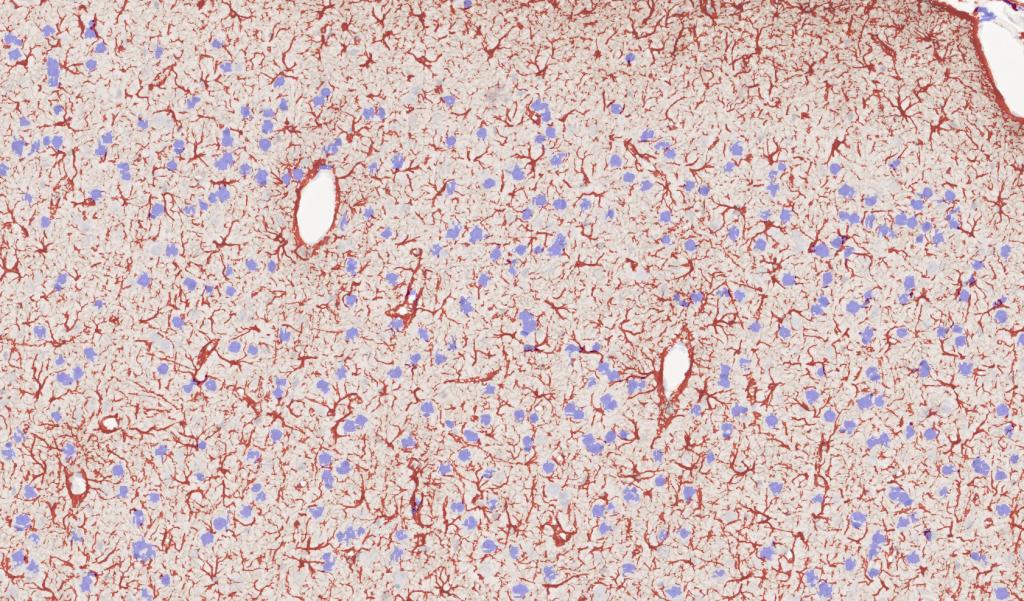
Figure 3. Rat brain exposed to severe traumatic brain injury (TBI) via mechanical insult; tissue section labeled with IHC for GFAP, drag slider to visualize segmentation depicting astrocytes in red, and neurons in blue.
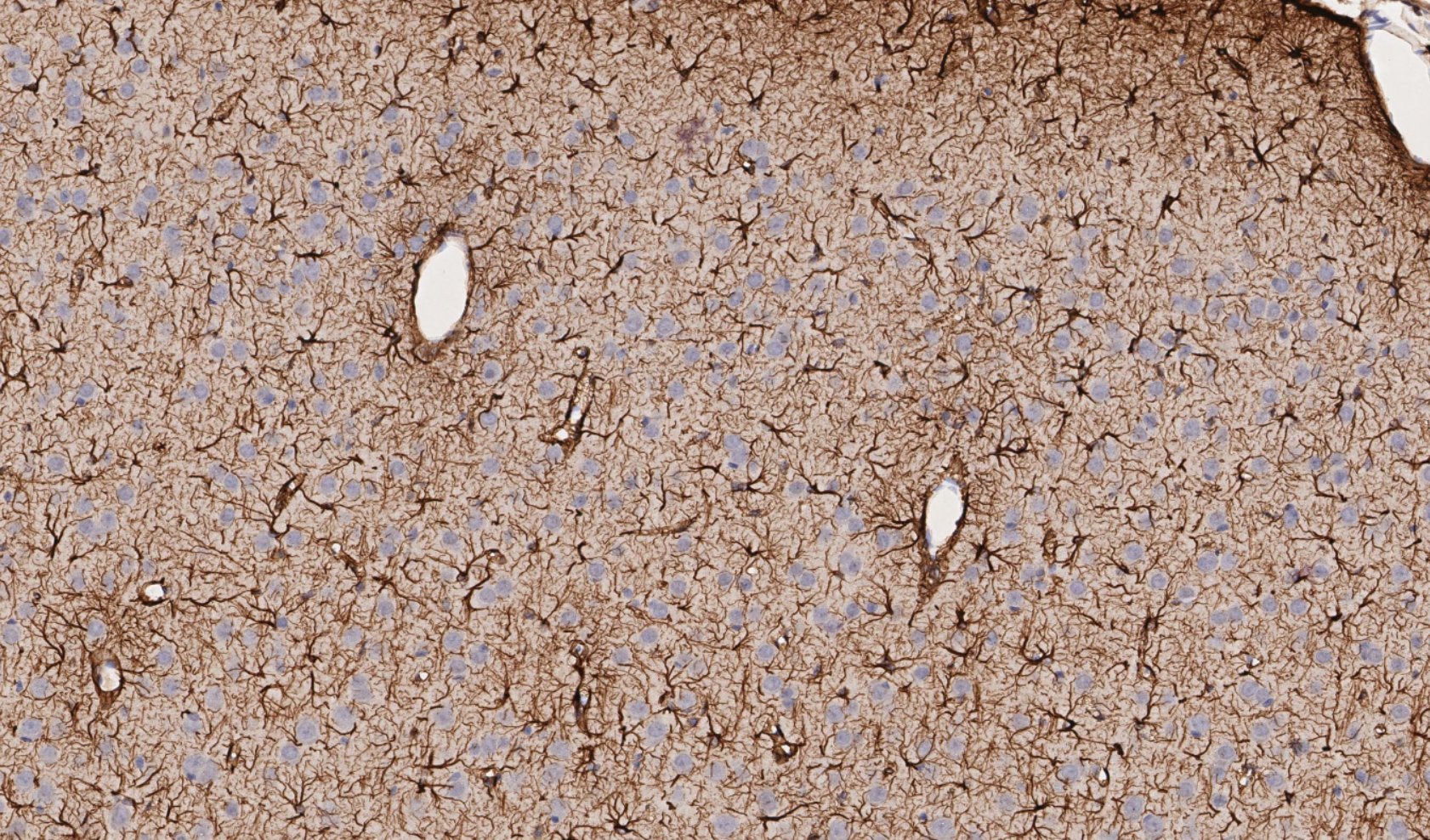
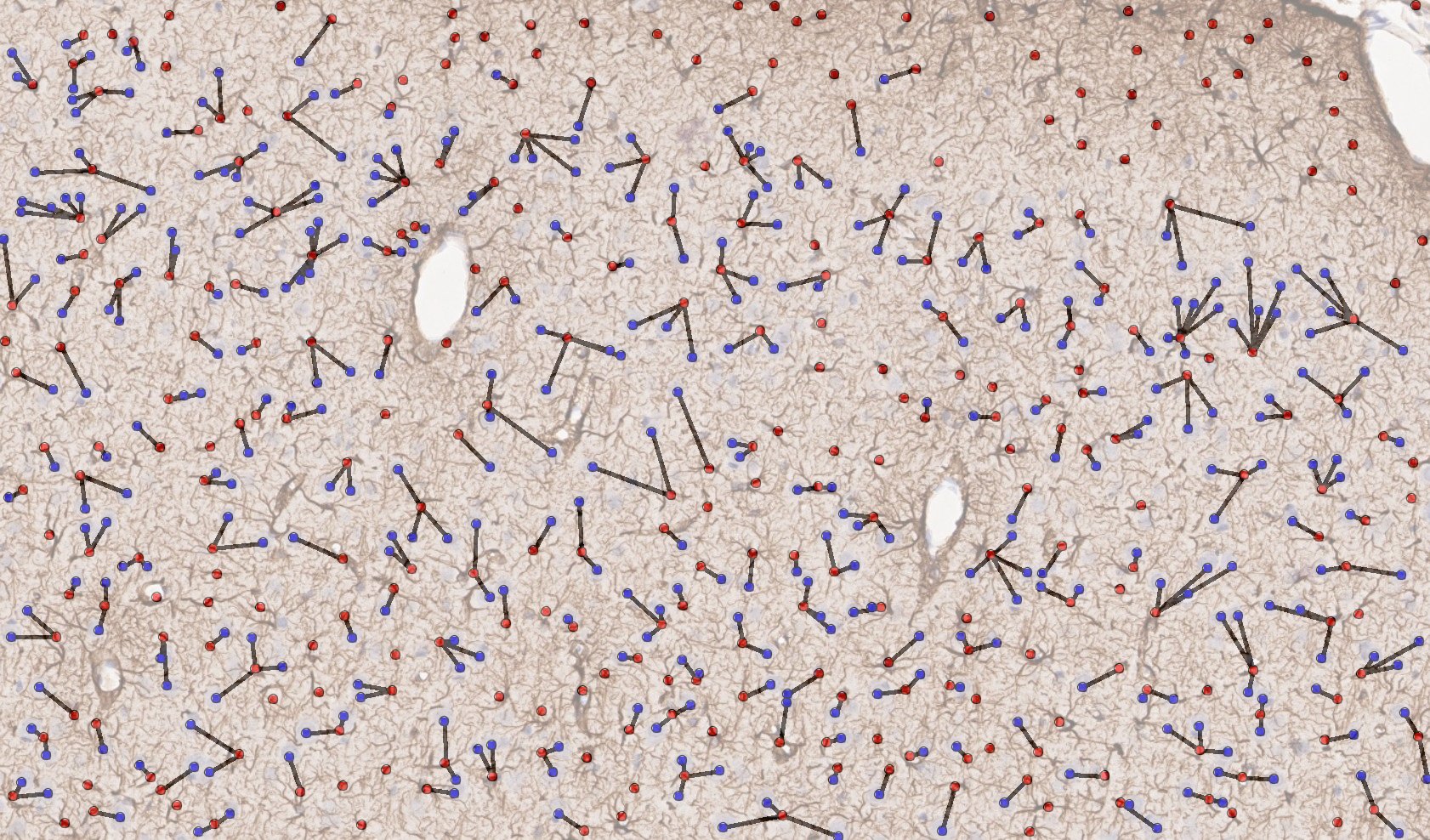
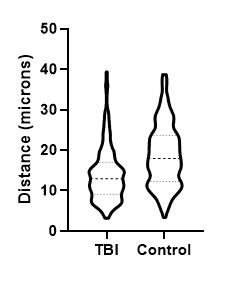
Figure 4. Mechanically induced TBI in rat brain; representative region of IHC tissue section labeled with GFAP and nuclei labeled with hematoxylin, drag slider to reveal graphical representation of nearest neighbor analysis used to assess neuron-astrocyte distances. Violin-plot depicts distribution of neuron-astrocyte distances in TBI (reactive astrocytes) and control brain tissue. There was a statistically significant decrease in the average distance between neurons and reactive astrocytes measured from GFAP labeled sections.


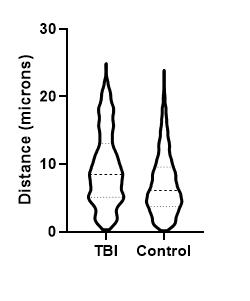
Figure 5. Mechanically induced TBI in rat brain; representative region of IHC tissue section labeled with GFAP and nuclei labeled with hematoxylin, drag slider to reveal graphical representation of nearest neighbor analysis used to assess astrocyte-blood vessel distances. Violin-plot depicts distribution of reactive astrocyte-blood vessel distances in TBI and control brain tissue. There was a statistically significant increase in the average distance between astrocytes and nearest blood vessel measured from GFAP labeled sections, likely due to increased GFAP expression following TBI due to astrocyte activation, leading to increased number of astrocytes detected by GFAP labeling distal from blood vessels.
References:
- Feldman, A.T., Wolfe, D. (June 2014). “Tissue Processing and Hematoxylin and Eosin Staining”. Histopathology. pp. 31-43.
- Distler, C., Dreher, Z., & Stone, J. (1991). Contact spacing among astrocytes in the central nervous system: an hypothesis of their structural role. Glia, 4(5), 484-494.
- Karve, I. P., Taylor, J. M., & Crack, P. J. (2016). The contribution of astrocytes and microglia to traumatic brain injury. British journal of pharmacology, 173(4), 692-702.