Overview
Evaluation of histological tissue sections plays a crucial role in disease diagnosis and understanding, as well as the assessment of therapies in drug discovery and development. Extracting quantitative data and analyzing image features from digital pathology slide images, which may not be visually discernible by a pathologist, offers opportunities for better disease modeling and potentially improved prediction of disease aggressiveness and patient outcomes.
With significant advancements in computer software and hardware, machine learning has emerged as a pivotal tool in various applications. In the field of bio-image analysis, major strides in computer vision and image processing techniques have greatly contributed to the analysis of histological images. Machine learning, specifically suited for image classification and categorization, has revolutionized the way we interpret and understand image content. At Visikol, we leverage the power of machine learning to provide cutting-edge classification services tailored to your specific needs.
Machine learning has emerged as a pivotal tool in image and data analysis, offering advanced techniques for classification and categorization. Our expert team harnesses the potential of machine learning algorithms to unlock hidden patterns, structures, and valuable information within your image or tabulated data.
We offer two main approaches for classification: supervised and unsupervised machine learning. Supervised machine learning involves training the algorithm using annotated data, allowing it to recognize important features and make accurate classifications. This method is particularly useful when you have labeled data and predefined categories. Our team utilizes various supervised machine learning techniques, including deep learning, neural networks, Random Forest, Naive Bayes, and Support Vector Machine algorithms, to provide accurate and reliable classifications.
Alternatively, unsupervised machine learning is employed when there are no predefined categories or labeled data. It allows for exploratory analysis and identification of underlying structures and patterns within your data. Using advanced unsupervised machine learning techniques such as clustering analysis and dimensionality reduction, we uncover meaningful insights from your image or tabulated data without prior knowledge of categories or labels.
Key Features of our Machine Learning Classification Services:
- Supervised Classification: We employ supervised machine learning techniques to classify histological sections into predefined categories based on expert annotations and known features.
- Unsupervised Classification: Our unsupervised machine learning approaches analyze image data to identify underlying structures and patterns without predefined categories or training sets.
- Advanced Algorithms: We leverage state-of-the-art machine learning algorithms and methodologies to ensure accurate and reliable classifications, tailored to your specific dataset.
- Customized Solutions: Our services are designed to meet the unique requirements of your project. We work closely with you to understand your goals and optimize our classification approach accordingly.
- Expert Analysis: Our team of experienced image analysts ensure high-quality interpretation and accurate classification of histological sections.
- Insightful Interpretation: We go beyond the classification itself and provide meaningful interpretations of the results, helping you gain valuable insights and make informed decisions.
Whether you need to classify images or analyze tabulated data, Visikol’s machine learning classification services empower you to unlock the hidden potential within your dataset. Our streamlined processes and advanced algorithms ensure accurate and efficient analysis, saving you time and effort.
Analysis Compatibilities and Deliverables
| File Formats | Images: TIFF, PNG, JPEG Tabular Data : CSV, XLSX |
| Image Submission | Hard Drive Visikol Cloud Sharing Client selected file sharing service |
| Data Delivery | Report Tabular data |
General Procedure
- Training data checked to make sure the quality is good for training.
- The best model for the classification task is determined through a preliminary evaluation.
- The optimal model is trained with the data provided and precision, recall, and accuracy are used to determine model performance.
- The trained model is validated against a separate testing set to ensure robustness.
- A report is sent to the client about the model development.
- Next steps to be determined with client for use of model or implementation.
Representative Data
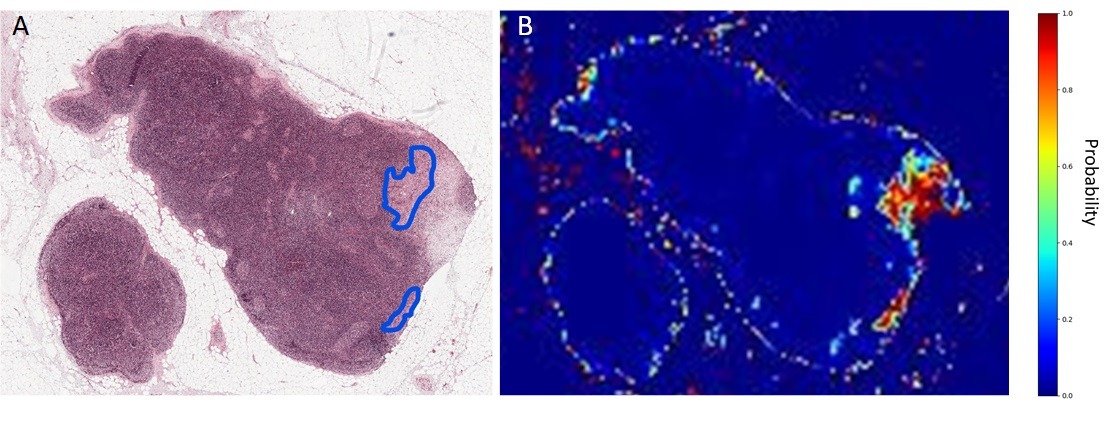
Figure 1. Results of supervised machine learning classification of potential tumor regions in sentinel lymph node section.
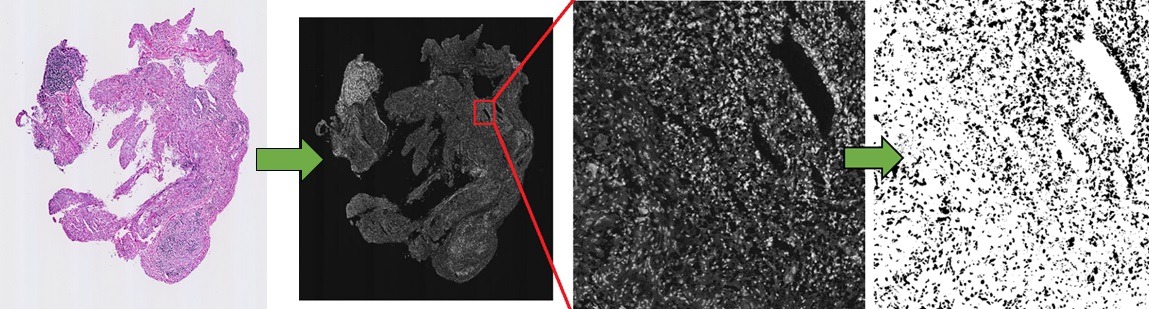
Figure 2. Representation of process of extraction of features for use in unsupervised machine learning classification. In this example, the nuclear channel is extracted and segmented to measure nuclear morphology, and the resultant data is utilized to classify tissue samples according to variations in nuclear morphology.
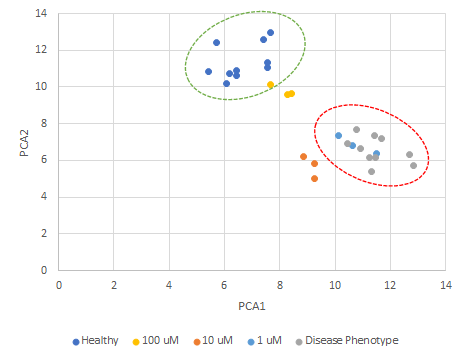
Figure 4. Result of principal component analysis on the quantitative measurements of nuclear morphology from a set of histological samples showing the shift in phenotype from diseased to healthy upon drug treatment.

