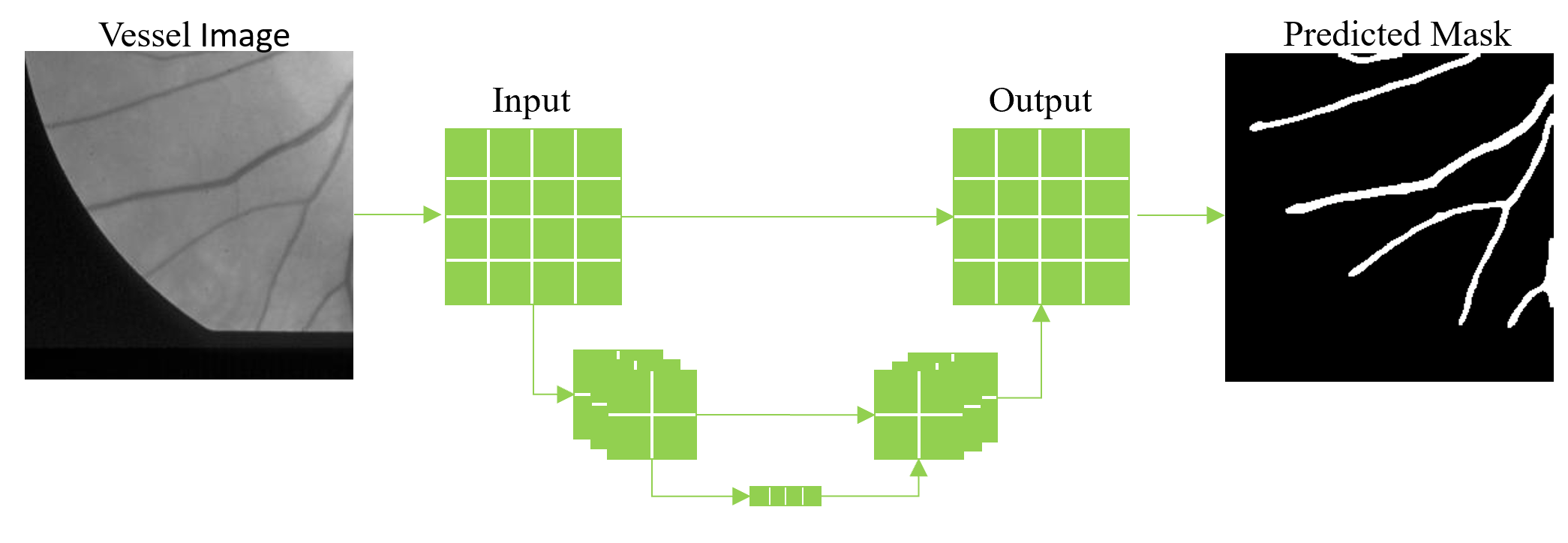
Blood vessel analysis has become an important aspect among many disciplines resulting in automated vessel segmentation algorithms to be a crucial step in scientist’s and physician’s workflow. Manual segmentation requires a high level of expertise and is very time consuming creating a need for accurate, reliable segmentation algorithms. A variety of methods already exist and are often categorized into different classes such as vessel enhancement, machine learning, deformable models, and tracking.1 In this blog post we discuss a supervised deep learning approach using publicly available datasets with ground truth annotations provided by experts in the field.
Semantic segmentation differs from instance segmentation in that semantic segmentation is not concerned with differentiating between different objects belonging to the same class. The main goal of semantic segmentation is to partition pixels in the input image into their retrospective classes provided by the ground truth data. For vessel segmentation it is important to realize that instance segmentation-based models, which are highly complex, are not necessary for the given task. In our 3Screen™ software, we utilize a fully convolutional network architecture called U-Net2 which is highly popular among the biomedical and life science community for several reasons: 1) It has proved especially effective on biomedical datasets, 2) It performs well on small datasets, and 3) It is simplistic to understand based on its elegant encoder-decoder architecture. The architecture resembles a ‘U’ because the input image travels down the ‘U’ during the encoding phase where it is passed through a series of convolutional and pooling layers, effectively down-sampling the image in the X-Y dimensions and increasing the size in the Z dimension. Next, the intermediate data travels up the ‘U’ during the decoding phase and is up-sampled in the X-Y dimension with an operation called “deconvolution” with the same amount of filters as the encoding phase. It is considered fully convolutional because the decoding layers are connected to both the previous decoding layer and a corresponding encoding layer which helps preserve spatial information due to the down-sampling which occurs in the encoding phase.
Below we show applying our 3Screen™ software to publicly available retinal datasets from the DRIVE3 and STARE4 database to train a convolutional neural network (CNN) to perform vessel segmentation on images acquired using color fundus photography (CFP). We evaluated the trained CNN on the test images provided by the DRIVE database which is a popular test set for evaluating vessel segmentation performance. Both datasets contain only a small number of retinal images (40 total) and are quite large (DRIVE 565×584; STARE 605×700). Therefore, we split each full-size image into multiple smaller images called “patches” with dimensions 256×256. By doing this we are able to extract hundreds of unique patches from each full-size image which increases the number of training samples drastically. As mentioned previously, we use the U-Net architecture to train a CNN to perform vessel segmentation. To perform inference on a full-size image, we deconstruct the image into multiple 256×256 patches to perform segmentation and then reconstruct the segmented patches into a full-size segmentation mask. On the test set provided by the DRIVE database, we achieved an accuracy of 96.5%, sensitivity 78.4%, specificity 98.2%, and Dice similarity coefficient 0.797.
Compared to another successful segmentation algorithms involving the Hessian matrix, this deep learning approach outperforms the Hessian algorithm with respect to accuracy, sensitivity, specificity, and the Dice similarity coefficient on the DRIVE test set. This is largely because the CNN has learned to discriminate against noise very well while also handling instances of variable brightness and contrast. The Hessian approach often struggles in very bright spots with curvature such as near the optic nerve or the border surrounding the eye, producing false positives. Using deep learning we are able to tackle some of these common problems and produce a model that is highly accurate and generalized for vessel segmentation in retinal images.
References
- Moccia, Sara, et al. “Blood vessel segmentation algorithms—Review of methods, datasets and evaluation metrics.” Computer methods and programs in biomedicine158 (2018): 71-91.
- Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. “U-net: Convolutional networks for biomedical image segmentation.” International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
- Staal, Joes, et al. “Ridge-based vessel segmentation in color images of the retina.” IEEE transactions on medical imaging4 (2004): 501-509.
- Hoover, Adam, Valentina Kouznetsova, and Michael Goldbaum. “Locating blood vessels in retinal images by piece-wise threshold probing of a matched filter response.” Proceedings of the AMIA Symposium. American Medical Informatics Association, 1998.