Visikol leverages its expertise in the field of advanced imaging and image analysis – with special emphasis on 3D – to execute service projects for Clients interested in analyzing organs, tissues, and tissue sections in greater qualitative and quantitative detail.
One of the most common methods used in histopathological analysis for research or diagnostic purposes is to cut thin sections from a paraffin block of formalin fixed tissue to and stain them with hematoxylin and eosin, also known simply as H&E. Both are chromogenic stains that label the nuclei a blueish-purple color and the cytoplasm pink, respectively, yielding a contrast of cellular features that is easily interpretable. As a result, the morphology of the tissue can be quickly and accurately examined by a pathologist using transmitted light microscopy. While this method is robust and time-tested, it still has a number of limitations and shortcomings. These include the fact that microtomy is required to generate thin sections, limiting the amount of tissue being sampled (often <1% of sample), it is destructive and non-reversible, meaning once the H&E-stained section is generated, it is no longer possible to conduct other labeling analyses, and it is subject to significant non-uniformity in staining quality, making direct comparisons between samples generated in different labs very difficult. In addition, the process of going from a wet fixed tissue specimen to an H&E stained slide can take quite a substantial amount of processing time and steps, and the resulting section can ultimately only be examined with brightfield microscopy.
To circumvent some of the pitfalls in traditional H&E, fluorescent labeling – combined with tissue clearing and confocal microscopy – can be used to generate images of a consistent quality in a much more streamlined way, with the added benefit of being able to optically section tissues in order to generate 3D image stacks. However, interpretation of fluorescently labeled tissues is not as straightforward, even when the only markers present are for nuclei and cytoplasm. Thus, a method of deriving “virtual H&E” images is implemented, in which the signal from each of the fluorescent channels is convolved and mapped in a specific manner to the RGB color space in order to render an image representative of the true nuclear and cytoplasmic morphology that would be in the equivalent H&E image.
For example, this method can be applied to breast cancer tissue specimens obtained from formalin-fixed and paraffin embedded (FFPE) blocks of tissue that are first deparaffinized, and subsequently stained with fluorescent labels for nuclei (DAPI) and cytoplasm (eosin) – see Figure 1. The methodology can be generally applied and is useful in a number of applications ranging from qualitative morphological evaluation of tissue specimens, to quantitative analysis of nuclear counts in metastases of cancerous tumor samples. The ability to perform 3D H&E on whole tissues and sections derived from fluorescent labeling opens the door for a wide range of applications in histopathological analysis that would otherwise be unavailable with the traditional version.
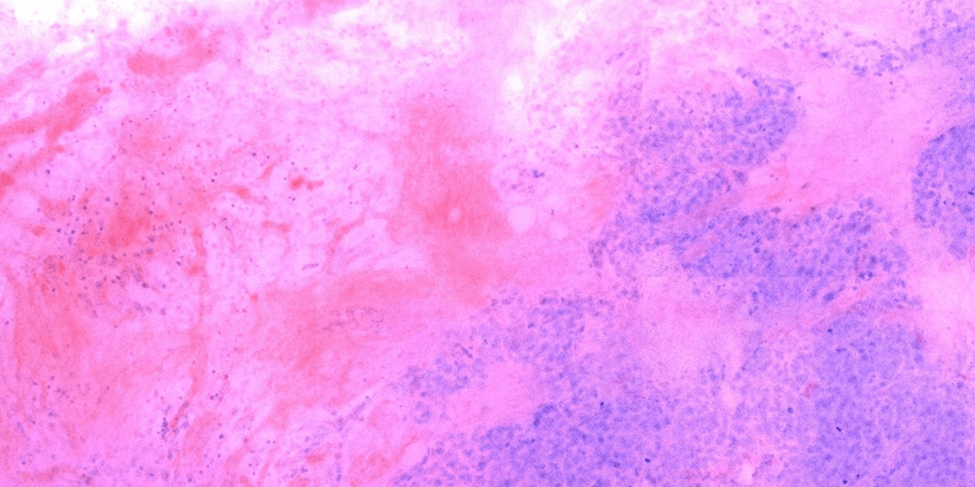
Figure 1. Virtual H&E image of tissue derived from an optical section taken from a thick piece of FFPE breast cancer lumpectomy tissue.

