Plasma protein binding refers to the noncovalent interaction of drugs with plasma proteins. Once the drug is in the bloodstream, it will exist in free and plasma protein-bound forms. An equilibrium will exist between the two forms, and it is only the unbound drug molecules that exert the pharmacological/toxicological effect. This is referred to as the “free drug hypothesis” Plasma contains several carrier proteins that facilitate the circulation of endogenous and exogenous molecules. Albumin is the most abundant protein in human plasma (~50%), which help maintain the osmotic pressure and the pH in the bloodstream. Examples of molecules that bind to albumin include peptides, fatty acids, and small-molecule drugs. The second most important plasma protein with drug-binding properties is an α1-acid glycoprotein (AAG).
The plasma protein binding assay is an important aspect of the in vitro ADME (absorption, distribution, metabolism, and excretion) field, which helps design and structure improvement of a drug candidate. A drug-like property includes solubility, permeability, metabolic stability, and protein binding. Traditionally, plasma protein binding assays are performed using 3H- or 14C-labeled compounds, which requires additional synthetic steps. However, with recent progress in automation, mass spec, and developing 96-well formats, LC-MS/MS is more routinely used and is considered the golden standard in plasma protein binding assays.
Experimentally, the tested compound is spiked into a plasma to a certain concentration (1-5 µM) and allowed to incubate at 37 oC via different approaches such as ultrafiltration and equilibrium dialysis. For ultrafiltration experiments, an aliquot of the compound-spiked plasma is transferred to an individual well, which has a filtration membrane at the bottom of the well with a 10,000 Da molecular weight cutoff. After centrifugation, the unbound analyte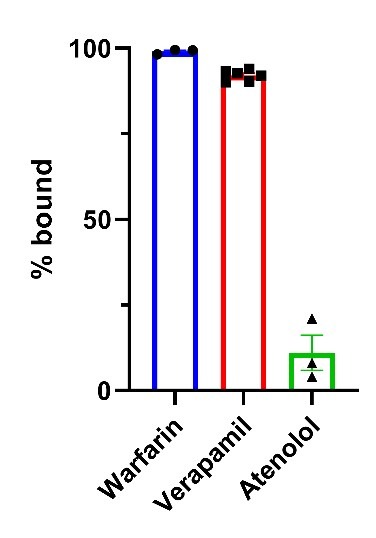 will move through the filter and be quantitively quantified, and the percentage of the unbound to the bound drug is calculated. Equilibrium dialysis is more popular in plasma protein binding assays and is considered the best choice for drug candidates. Briefly, the plasma-spiked compound is allowed to equilibrate with a phosphate buffer at 37 oC through a dialysis membrane for around 4 hours. LC-MS/MS measurements allowed the quantification of the concentration of the tested compound in both chambers; hence, the % bound to plasma proteins is calculated.
will move through the filter and be quantitively quantified, and the percentage of the unbound to the bound drug is calculated. Equilibrium dialysis is more popular in plasma protein binding assays and is considered the best choice for drug candidates. Briefly, the plasma-spiked compound is allowed to equilibrate with a phosphate buffer at 37 oC through a dialysis membrane for around 4 hours. LC-MS/MS measurements allowed the quantification of the concentration of the tested compound in both chambers; hence, the % bound to plasma proteins is calculated.
At Visikol, we used the equilibrium dialysis approach to evaluate the plasma protein binding affinity of three known drugs, warfarin, verapamil, and atenolol. Warfarin is a drug used to prevent blood clots. However, it has a narrow therapeutic window due to its high plasma protein bounding ability (~ 99% plasma protein bound). Our data confirmed warfarin’s high plasma protein binding ability and showed 99.5% in vitro plasma protein bound affinity. Atenolol, a beta blocker used to treat high blood pressure, showed the opposite profile with only 1.1 % in vitro plasma protein binding ability.
If you are interested in working with Visikol on your in vitro ADME project, please reach out to our team today.
