DNA methylation is a critical epigenetic modification that controls gene expression and genome stability, which has oncogenic implications during cancer initiation. DNA methylation occurs mainly on the CpG islands, which are mainly enriched at gene promoters. Hypermethylated promoters have been shown to silence the corresponding gene expression, while hypomethylation has the opposite effect. Interestingly, cancer is associated with mixed DNA methylation dysregulation, which includes a genome-wide hypomethylation and hypermethylation associated with the silencing of vital tumor-suppressor genes.[i] Retrieving gene-methylation patterns is an important biomarker for early cancer prediction and other diseases.
Whole-genome bisulfite sequencing is a well-established protocol for detecting methylated cytosines in genomic DNA. This protocol uses bisulfite-based deamination of unmethylated cytosine into uracil bases, while the methylated cytosine bases are unaffected. During sequencing, uracil bases are deaminated into thymidine, while methylated cytosines, which resisted the deamination step, are sequenced into cytosines (Figure 1A). Site-specific quantification of the generated cytosines provides information on the methylation status of the selected DNA region. One of the tools that can be used to retrieve such information is the LC-MS/MS.
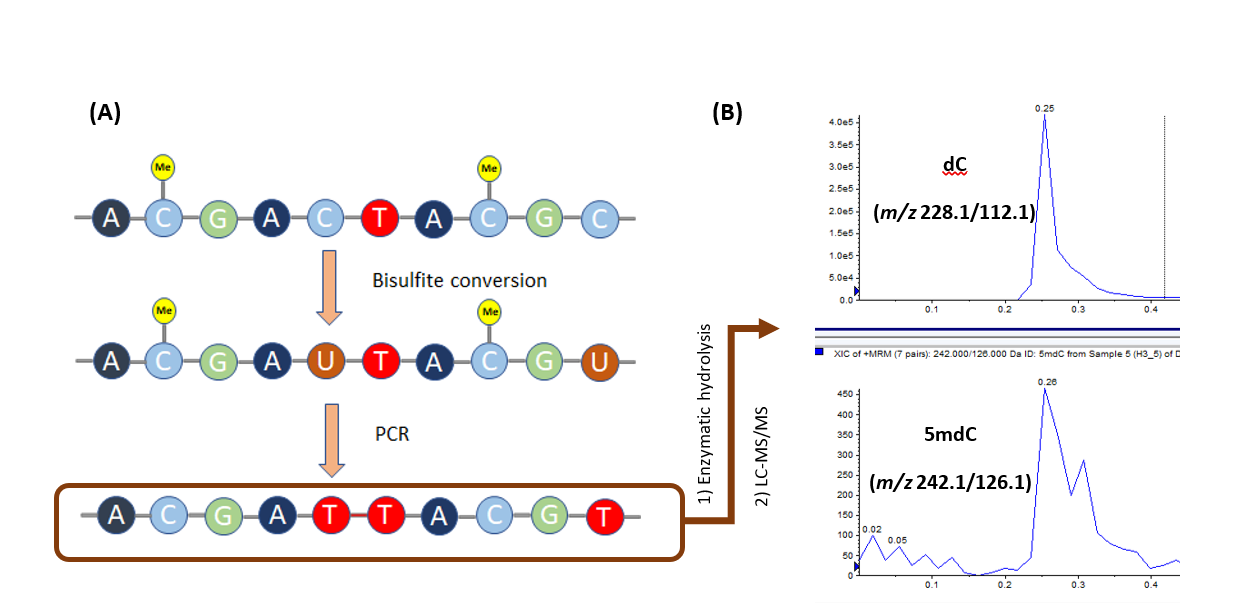
Figure 1. A) Overview of the bisulfite conversion-PCR sequence. B) LC-MS/MS selected base peak chromatogram of dC (top) at m/z 228.1/112.1 and 5mdC at m/z 242.1/126.1 (bottom) obtained from the enzymatic hydrolysis of 150 ng of the P53-amplified amplicon via DNA Degradase Plus (Zymo Research, E2021). As expected, 5mdC is not detected, which was lost during the bisulfite conversion step. Instead, a strong peak of dC was present in the P53 amplicon which is equivalent to the original methylation status of that gene.
Visikol has recently developed a sensitive bioanalytical method for the quantification of global DNA methylation by LC-MS/MS, which is based on the enzymatic hydrolysis of the genomic DNA and the simultaneous quantification of the corresponding nucleotides, 5-methyl-2’-deoxycytidine (5mdC), and 2’-deoxycytidine (dC), using the QTRAP 4000 mass spectrometer. This method was used to measure the changes in the global DNA methylation status of two cancer cell lines, A549 (human lung carcinoma) and HCT116 (human colorectal carcinoma), following the treatment with 5 µM of the DNA methyltransferase inhibitor 5-azacytidine (5-AZA). Using the DNA samples extracted from the 5-AZA-treated HCT116 cancer cell line, we extended the applicability of our method to quantify DNA methylation of the promoter region of the tumor suppressor gene, the p53 gene. Briefly, DNA samples were bisulfite converted, which then underwent a PCR amplification based on the following primers: ATTGTTTAGTTTTGTGTTAGGAGTTT, forward strand, and TCAATCAAAAACTTACCCAATCC for the reverse strand. The amplicons were then purified, subjected to our optimized enzymatic hydrolysis protocol, and finally analyzed by LC-MS/MS (Figure 1). The molar concentration of generated cytosine bases in the PCR amplicon is equivalent to what is expected based on the original methylation status of that gene. As expected, the amplicon hydrolysate showed no deoxythymidine, dT at sites where dC was anticipated. This data showed that the methylation status of the p53 gene in the A549 and HCT116 cancer cells did not change after the treatment with 5-AZA for 72 hours. Interestingly, 5-AZA treatment induced aggregation of nuclear material and decrease in the cell organelles (Figure 2).
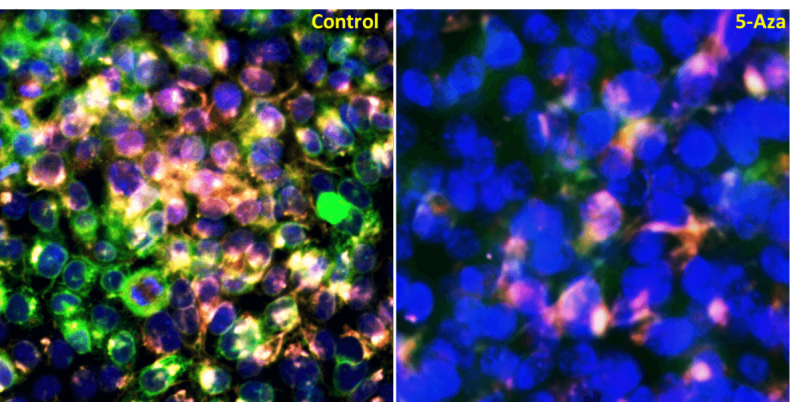
Figure 2. Cell painting of HCT116 cancer cells: Control and 5-AZA treated cells are stained with Hoechst (blue), Concanavalin A (green), SYTO14 (Magenta), WGA (yellow), and Mito tracker (deep red). 5-Aza shows aggregation of nuclear material and inhibition of cellular components.
If you are working on an epigenetic drug discovery project and need help quantifying your global DNA methylation or a gene-specific methylation, please reach out to our team to explore your epigenetic needs.
Reference: [i] Skvortsova, K.; Stirzaker, C.; Taberlay, P. The DNA methylation landscape in cancer. Essays Biochem., 2019, 6, 797-811.
