Visikol is the leading provider of multiplex tissue imaging services in the marketplace and provides clients with the broadest suite of approaches to multi-parametric imaging. Depending on a researcher’s specific research question, Visikol has a unique solution which is capable of balancing cost, throughput, markers and quality.
IMC allows researchers to visualize up to 40+ markers on a single slide while providing minimal background noise which means very high sensitivity imaging. Additionally, as IMC is based on the quantification of non-naturally occurring metals, so there is no Autofluorescence and very little background noise. The readouts for each antibody are a direct quantification of that antibody’s respective epitope, whereas with fluorescent imaging this type of quantification can be challenging.
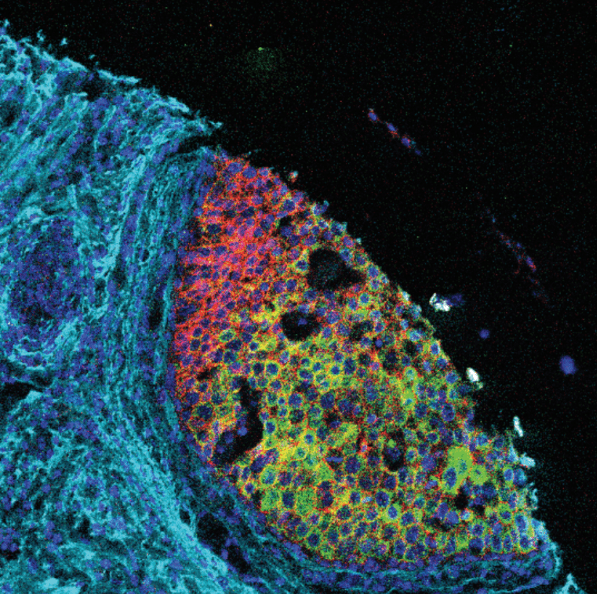
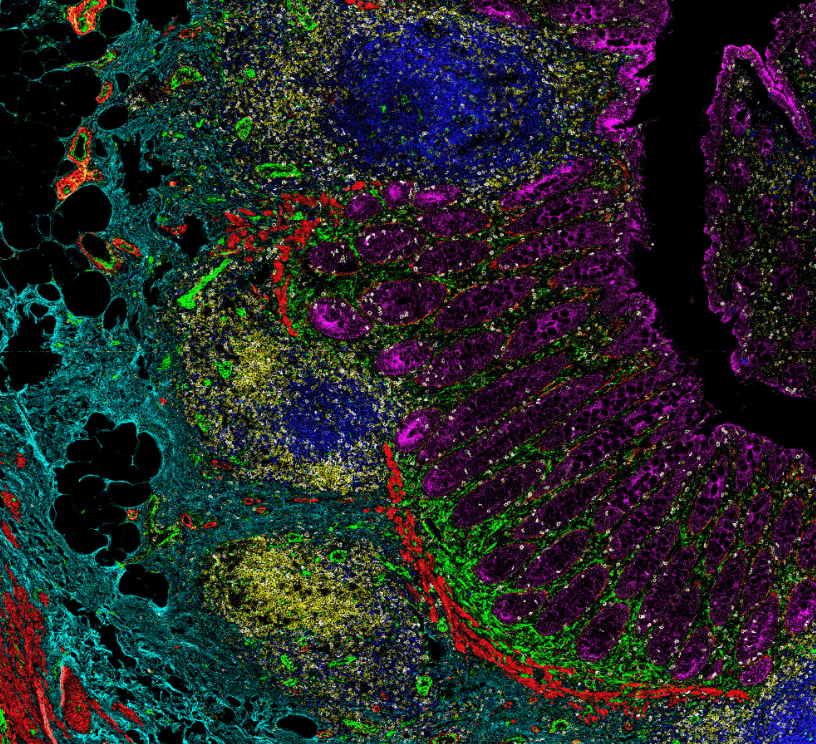
Advantages of Imaging Mass Cytometry
- Subcellular Resolution: Imaging quality and resolution equivalent to 10X optical microscopy for effective cell segmentation.
- No Autofluorescence: Using non-naturally-occurring metals for tagging antibodies limits potential pitfalls that can impact reproducibility and/or data quality.
- Rapid Labeling and Turnaround: Unlike fluorescent multiplexing, all of the labels can be used simultaneously which reduces the need for multiple rounds of labeling and tedious processing.
- Proven: Over 200 IMC publications provides confidence in results
Example 17-plex IMC Panel
An example 17-plex panel for IMC combines 3 panels together to assess the complexities of
the immune landscape within the tumor microenvironment.
Human Tissue Architecture Panel
Enables staining of tissue architecture,
including stroma, epithelial tissue,
and vasculature.
- Alpha-smooth muscle actin
- Collage type I
- E-cadherin
- Histone H3
- Vimentin
Human Tumor Infiltrating Lymphocytes Panel
Enables staining of epithelial cells; phenotyping of T cells (helper, cytotoxic, memory, and regulatory), B cells, and monocytes/macrophages; and identification of potential CD68+ tumor cells.
- CD20
- CD3
- CD4
- CD45RO
- CD68
- CD8a
- FoxP3
- Pan-keratin
Human Immune
Activation Panel
Allows for the phenotyping of
activated T cells.
- Granzyme B
- Ki-67
- PD-1
- PD-L1
For a list of antibodies validated for use in IMC using Maxpar® OnDemand Antibodies click the button below.
