HepaRG™ NP 3D model features: Relevant cell subtypes present
The HepaRG™ NP 3D liver models feature liver cell subtypes in proportions found in the native human liver. These include hepatocytes, hepatic biliary cells, liver endothelial cells, Kuppfer cells, and stellate cells, which are relevant not only to normal physiologic liver function but also to the progression of liver injury and disease.
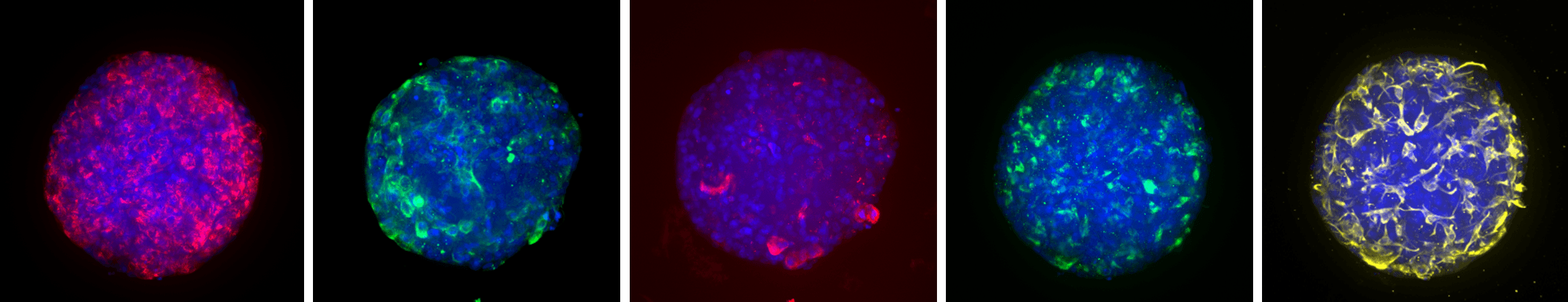
HepaRG™ NP 3D model labeled with DAPI (blue) and, from left to right, albumin (red; hepatocytes), pan-cytokeratin (green; biliary cells), CD31 (red; liver endothelial cells), CD68 (green; Kupffer cells), vimentin (yellow; stellate cells).
HepaRG™ NP 3D model features: Other relevant features
The HepaRG™ NP 3D liver models feature a number of other features relevant to normal liver functionality and to the progression of liver disease and toxicity. These include glycogen storage, hepatobiliary transporter expression, and drug metabolizing enzyme expression and inducibility among other features.
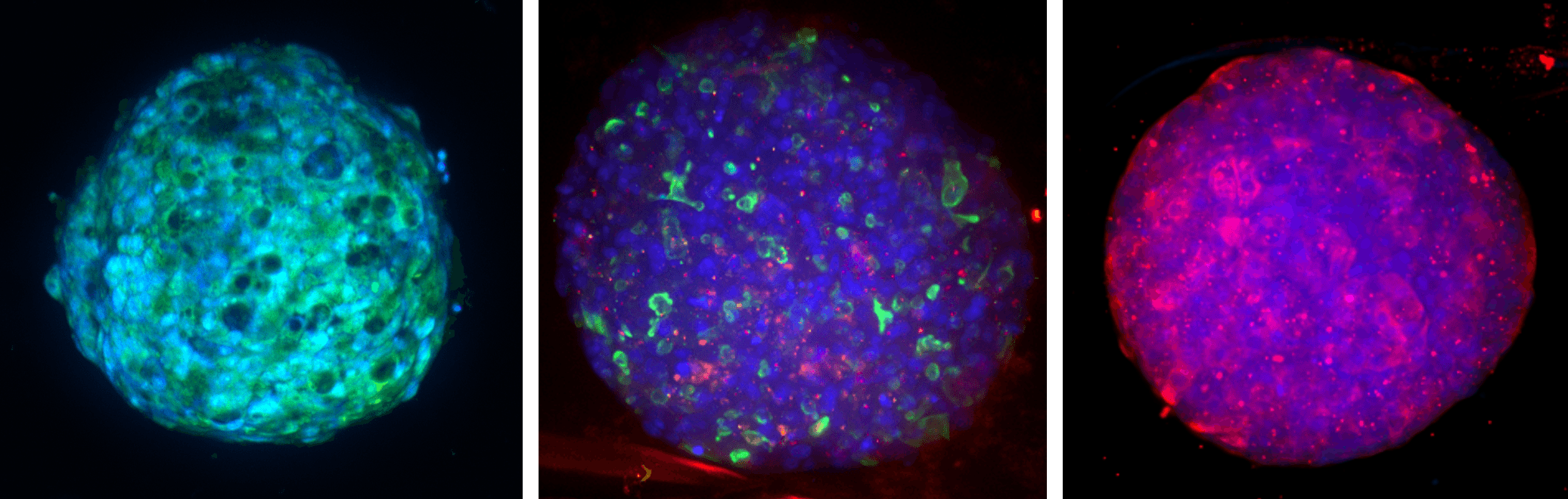
HepaRG™ NP 3D model labeled with DAPI (blue) and, from left to right, PAS (green; hepatocytes), hepatobiliary transporters (MRPII in green, MDRI in red), Cyp3A4 (red).
HepaRG™ NP 3D model capabilities: Drug induced liver injury (DILI)
Evaluating liver toxicity is crucial to many drug discovery and development initiatives, and the ability to do so in vitro can increase throughput and thus reduce the costs and risks associated with bringing a new drug to market. The HepaRG™ NP 3D model features the ability to recapitulate a number of modes of liver toxicity including non-cholestatic, cholestatic, and immune-mediated mechanisms. For more information on evaluating DILI, visit our hepatotoxicity assay page.
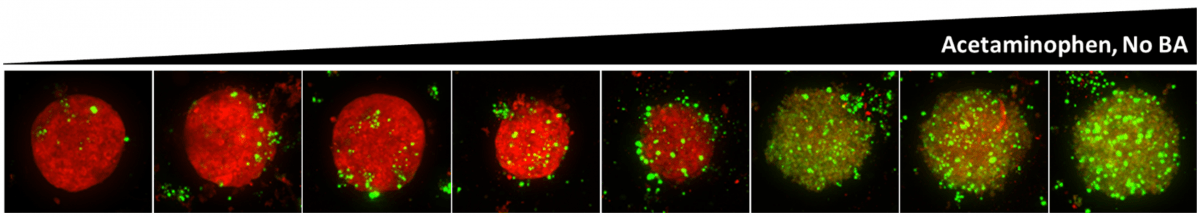
Maximum Z-projection of Z-stack obtained from High Content Imaging of OpenLiver HepaRG/NP 3D cell models treated with acetaminophen. Red: Mitochondrial function probe; Green: cell viability probe
HepaRG™ NP 3D model capabilities: Fibrosis
The HepaRG™ NP 3D also feature the ability to recapitulate a number of liver disease pathologies. Whether evaluating the disease-inducing liability of a compound or exploring targets to exploit for therapeutic amelioration of disease state, the HepaRG™ NP 3D is capable of recapitulating disease features such hepatocellular lipid accumulation, nonparenchymal cell phenotype shift, and collagen matrix deposition. For more information on these assays, visit our assay pages on steatosis and fibrosis.
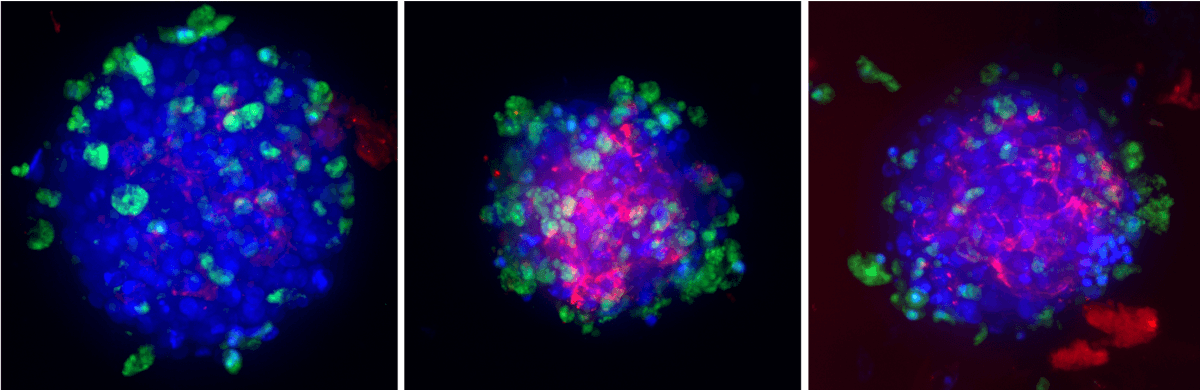
Visikol HepaRG/NP 3D spheroids were pretreated with either vehicle or an ALK-5 inhibitor for 1 h prior to inducing fibrosis with 100 nM TGF-β for 48 h. DAPI in blue, viability indicator in green, pan collagen in red.
Cell culture protocol
The Visikol HepaRG™ NP 3D model is generated from the co-culture of HepaRG cells with single-donor mixed nonparenchymal cells (NPCs). To generate these models, cells are thawed, washed with complete media (HepaRG General Purpose Medium + 1X anti-anti), and immediately resuspended in complete media at concentrations sufficient for plating 2×103 total cells per well of either a Corning 96 or 384 Well Ultra-Low Attachment Treated Spheroid Microplate at a 60:40 ratio of HepaRG:NPCs. Spheroids begin to aggregate within 24 h of initial plating, after which media is changed on day 3.
